Note
Go to the end to download the full example code.
Examples of the KSpaceFilter class¶
- Authors:
Michele Ceriotti @ceriottm
This notebook demonstrates the use of the KSpaceFilter
class to transform a density by applying a scalar filter in reciprocal space.
The class supports many different use cases, and can be reused several times if the filter or the mesh size don’t change.
from time import time
import ase
import chemiscope
import numpy as np
import torch
from matplotlib import pyplot as plt
import torchpme
device = "cpu"
dtype = torch.float64
Demonstrates the application of a k-space filter¶
We define a fairly rugged function on a mesh, and apply a smoothening filter. We start
creating a grid (we use a MeshInterpolator object for simplicity to generate
the grid with the right shape) and computing a sharp Gaussian field in the \(xy\)
plane.
cell = torch.eye(3, dtype=dtype, device=device) * 6.0
ns_mesh = torch.tensor([9, 9, 9])
interpolator = torchpme.lib.MeshInterpolator(
cell, ns_mesh, interpolation_nodes=2, method="P3M"
)
xyz_mesh = interpolator.get_mesh_xyz()
mesh_value = (
np.exp(-4 * ((cell[0, 0] / 2 - xyz_mesh) ** 2)[..., :2].sum(axis=-1))
).reshape(1, *xyz_mesh.shape[:-1])
/home/runner/work/torch-pme/torch-pme/examples/04-kspace-demo.py:49: DeprecationWarning: __array_wrap__ must accept context and return_scalar arguments (positionally) in the future. (Deprecated NumPy 2.0)
np.exp(-4 * ((cell[0, 0] / 2 - xyz_mesh) ** 2)[..., :2].sum(axis=-1))
To define and apply a Gaussian smearing filter, we first define the convolution kernel
that must be applied in the Fourier domain, and then use it as a parameter of the
filter class. The application of the filter requires simply a call to
lib.KSpaceKernel.compute().
# This is the filter function. NB it is applied
# to the *squared k vector norm*
class GaussianSmearingKernel(torchpme.lib.KSpaceKernel):
def __init__(self, sigma2: float):
self._sigma2 = sigma2
def kernel_from_k_sq(self, k_sq):
return torch.exp(-k_sq * self._sigma2 * 0.5)
# This is the filter
kernel_filter = torchpme.lib.KSpaceFilter(
cell, ns_mesh, kernel=GaussianSmearingKernel(sigma2=1.0)
)
# Apply the filter to the mesh
mesh_filtered = kernel_filter.forward(mesh_value)
Visualize the effect of the filter¶
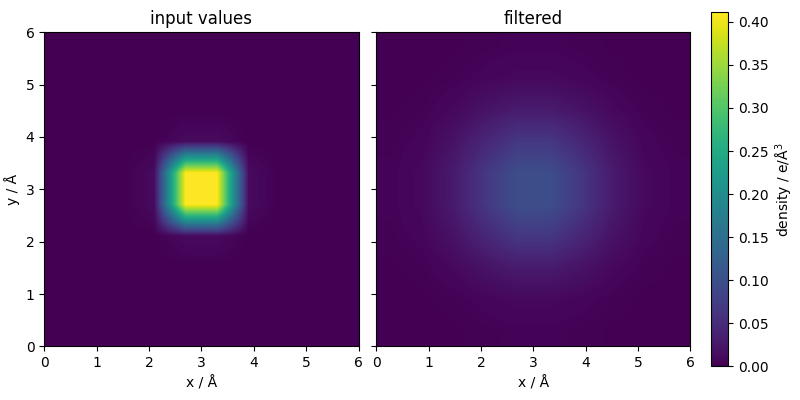
The Gaussian smearing enacted by the filter can be easily visualized taking a slice of the mesh (we make it periodic for a cleaner view)
fig, ax = plt.subplots(
1, 2, figsize=(8, 4), sharey=True, sharex=True, constrained_layout=True
)
mesh_extent = [
0,
cell[0, 0],
0,
cell[1, 1],
]
z_plot = mesh_value[0, :, :, 0].detach().numpy()
z_plot = np.vstack([z_plot, z_plot[0, :]]) # Add first row at the bottom
z_plot = np.hstack(
[z_plot, z_plot[:, 0].reshape(-1, 1)]
) # Add first column at the right
z_min, z_max = (z_plot.min(), z_plot.max())
cf = ax[0].imshow(
z_plot,
extent=mesh_extent,
vmin=z_min,
vmax=z_max,
origin="lower",
interpolation="bilinear",
)
z_plot = mesh_filtered[0, :, :, 0].detach().numpy()
z_plot = np.vstack([z_plot, z_plot[0, :]]) # Add first row at the bottom
z_plot = np.hstack(
[z_plot, z_plot[:, 0].reshape(-1, 1)]
) # Add first column at the right
cf_fine = ax[1].imshow(
z_plot,
extent=mesh_extent,
vmin=z_min,
vmax=z_max,
origin="lower",
interpolation="bilinear",
)
ax[0].set_xlabel("x / Å")
ax[1].set_xlabel("x / Å")
ax[0].set_ylabel("y / Å")
ax[0].set_title(r"input values")
ax[1].set_title(r"filtered")
fig.colorbar(cf_fine, label=r"density / e/Å$^3$")
fig.show()
We can also show the filter in action in 3D, by creating a grid of dummy atoms corresponding to the mesh points, and colored according to the function value. Use the chemiscope option panel to switch between the sharp input and the filtered values.
dummy = ase.Atoms(
positions=xyz_mesh.reshape(-1, 3),
symbols="H" * len(xyz_mesh.reshape(-1, 3)),
cell=cell,
)
chemiscope.show(
frames=[dummy],
properties={
"input value": {
"target": "atom",
"values": mesh_value.detach().numpy().flatten(),
},
"filtered value": {
"target": "atom",
"values": mesh_filtered.detach().numpy().flatten(),
},
},
mode="structure",
settings=chemiscope.quick_settings(
structure_settings={
"unitCell": True,
"bonds": False,
"environments": {"activated": False},
"color": {
"property": "input value",
"transform": "linear",
"palette": "viridis",
},
}
),
environments=chemiscope.all_atomic_environments([dummy]),
)
/home/runner/work/torch-pme/torch-pme/examples/04-kspace-demo.py:147: UserWarning: `frames` argument is deprecated, use `structures` instead
chemiscope.show(
Adjustable and multi-channel filters¶
KSpaceFilter can also be applied for more complicated
use cases. For instance, one can apply multiple filters to multiple real-space mesh
channels, and use a torch.nn.Module-derived class to define an adjustable
kernel.
We initialize a three-channel mesh, with identical patterns along the three Cartesian directions
/home/runner/work/torch-pme/torch-pme/examples/04-kspace-demo.py:191: DeprecationWarning: __array_wrap__ must accept context and return_scalar arguments (positionally) in the future. (Deprecated NumPy 2.0)
np.exp(-4 * ((cell[0, 0] / 2 - xyz_mesh) ** 2)[..., :2].sum(axis=-1)),
/home/runner/work/torch-pme/torch-pme/examples/04-kspace-demo.py:192: DeprecationWarning: __array_wrap__ must accept context and return_scalar arguments (positionally) in the future. (Deprecated NumPy 2.0)
np.exp(-4 * ((cell[0, 0] / 2 - xyz_mesh) ** 2)[..., [0, 2]].sum(axis=-1)),
/home/runner/work/torch-pme/torch-pme/examples/04-kspace-demo.py:193: DeprecationWarning: __array_wrap__ must accept context and return_scalar arguments (positionally) in the future. (Deprecated NumPy 2.0)
np.exp(-4 * ((cell[0, 0] / 2 - xyz_mesh) ** 2)[..., 1:].sum(axis=-1)),
We also define a filter with three smearing parameters, corresponding to three channels
class MultiKernel(torchpme.lib.KSpaceKernel):
def __init__(self, sigma: torch.Tensor):
super().__init__()
self._sigma = sigma
def kernel_from_k_sq(self, k_sq):
return torch.stack([torch.exp(-k_sq * s**2 / 2) for s in self._sigma])
This can be used just as a simple filter
multi_kernel = MultiKernel(torch.tensor([0.25, 0.5, 1.0], dtype=dtype, device=device))
multi_kernel_filter = torchpme.lib.KSpaceFilter(cell, ns_mesh, kernel=multi_kernel)
multi_filtered = multi_kernel_filter.forward(multi_mesh)
When the parameters of the kernel or the cell are modified, one has to call
KSpaceFilter.update() before applying the filter
multi_kernel._sigma = torch.tensor([1.0, 0.5, 0.25])
multi_kernel_filter.update(cell)
multi_filtered_2 = multi_kernel_filter.forward(multi_mesh)
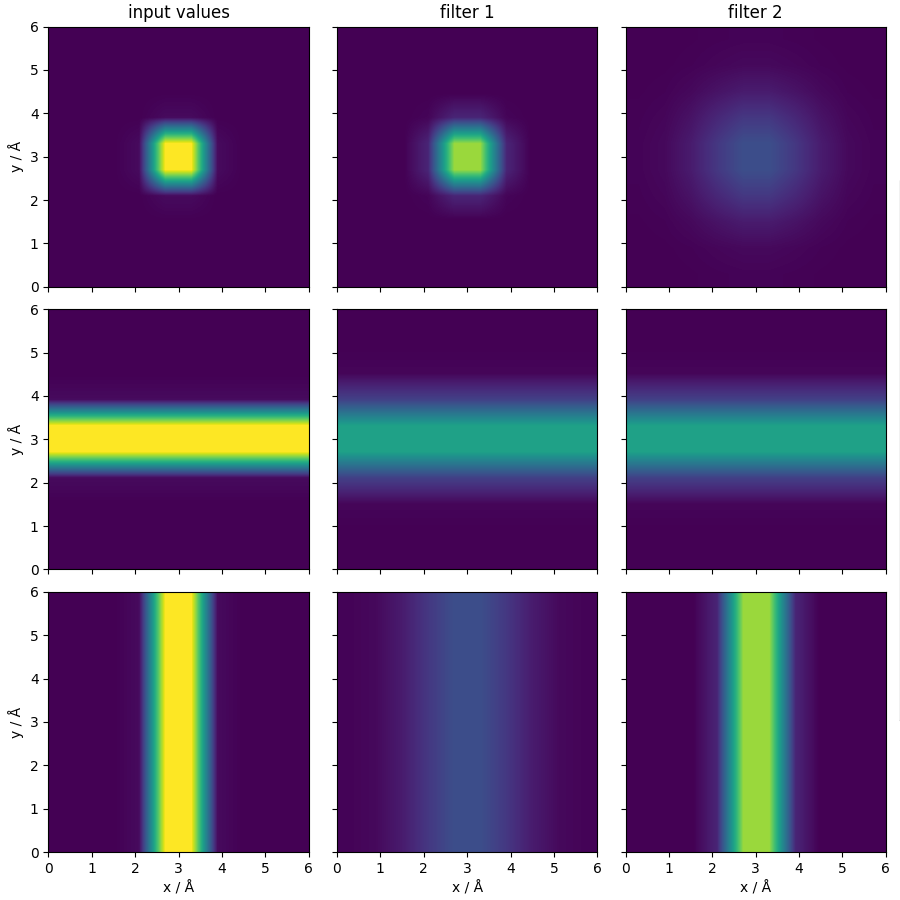
Visualize the application of the filters¶
The first colum shows the input function (the three channels contain the same function as in the previous example, but with different orientations, which explains the different appearence when sliced along the \(xy\) plane).
The second and third columns show the same three channels with the Gaussian smearings defined above.
fig, ax = plt.subplots(
3, 3, figsize=(9, 9), sharey=True, sharex=True, constrained_layout=True
)
# reuse the same mesh_extent given the mesh is cubic
mesh_extent = [
0,
cell[0, 0],
0,
cell[1, 1],
]
z_min, z_max = (mesh_value.flatten().min(), mesh_value.flatten().max())
cfs = []
for j, mesh_value in enumerate([multi_mesh, multi_filtered, multi_filtered_2]):
for i in range(3):
z_plot = mesh_value[i, :, :, 4].detach().numpy()
z_plot = np.vstack([z_plot, z_plot[0, :]]) # Add first row at the bottom
z_plot = np.hstack(
[z_plot, z_plot[:, 0].reshape(-1, 1)]
) # Add first column at the right
cf = ax[i, j].imshow(
z_plot,
extent=mesh_extent,
vmin=z_min,
vmax=z_max,
origin="lower",
interpolation="bilinear",
)
cfs.append(cf)
ax[j, 0].set_ylabel("y / Å")
ax[2, j].set_xlabel("x / Å")
ax[0, 0].set_title("input values")
ax[0, 1].set_title("filter 1")
ax[0, 2].set_title("filter 2")
cbar_ax = fig.add_axes(
[1.0, 0.2, 0.03, 0.6]
) # [left, bottom, width, height] in figure coordinates
# Add the colorbar to the new axes
fig.colorbar(cfs[0], cax=cbar_ax, orientation="vertical")
<matplotlib.colorbar.Colorbar object at 0x7f860072cb90>
Jit-ting of the k-space filter¶
The k-space filter can also be compiled to torch-script, for faster execution (the impact is marginal for this very simple case)
multi_filtered = multi_kernel_filter.forward(multi_mesh)
start = time()
for _i in range(100):
multi_filtered = multi_kernel_filter.forward(multi_mesh)
time_python = (time() - start) * 1e6 / 100
jitted_kernel_filter = torch.jit.script(multi_kernel_filter)
jit_filtered = jitted_kernel_filter.forward(multi_mesh)
start = time()
for _i in range(100):
jit_filtered = jitted_kernel_filter.forward(multi_mesh)
time_jit = (time() - start) * 1e6 / 100
print(f"Evaluation time:\nPytorch: {time_python}µs\nJitted: {time_jit}µs")
Evaluation time:
Pytorch: 95.40319442749023µs
Jitted: 115.45419692993164µs
Total running time of the script: (0 minutes 1.048 seconds)